Chlorarachniophytes
Ken-ichiro Ishida and Geoff I. McFadden

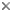
This tree diagram shows the relationships between several groups of organisms.
The root of the current tree connects the organisms featured in this tree to their containing group and the rest of the Tree of Life. The basal branching point in the tree represents the ancestor of the other groups in the tree. This ancestor diversified over time into several descendent subgroups, which are represented as internal nodes and terminal taxa to the right.

You can click on the root to travel down the Tree of Life all the way to the root of all Life, and you can click on the names of descendent subgroups to travel up the Tree of Life all the way to individual species.
For more information on ToL tree formatting, please see Interpreting the Tree or Classification. To learn more about phylogenetic trees, please visit our Phylogenetic Biology pages.
close boxIntroduction
If you find a green amoeba in your sea water sample, it may be a member of the chlorarachniophytes. The green color is caused by chloroplasts, which are permanently housed in the amoeba cell. Although the chloroplasts contain chlorophylls a and b, chlorarachniophytes are phylogenetically distinct from other chlorophyll b-containing eukaryotes (the chlorophytes and the euglenophytes), and constitute an independent phylum: Chlorarachniophyta, which was established only two decades ago (Hibberd and Norris 1984, Hibberd 1990). The group consists of four genera and five species, and there are also several species waiting to be named (Ishida et al. 1999, Ishida et al. 2000).
Although they are an obscure and little known group of algae, chlorarachniophytes have been a magnet for evolutionary biologists interested in the origin and the evolution of chloroplasts. This is because they are one of the groups that acquired their chloroplasts via a secondary endosymbiosis, in which a non-photosynthetic eukaryote engulfed an eukaryotic alga and retained it as a photosynthetic organelle (McFadden et al. 1994, McFadden et al. 1997). Indeed chlorarachniophytes are one of two groups (the other is cryptomonads) that provide watertight evidence for secondary chloroplast acquisition. Both chlorarachniophytes and cryptomonads have nucleomorphs, the reduced nuclei of the algal endosymbiont that gave rise to the chloroplasts (Ludwig and Gibbs 1989, McFadden et al. 1994, Gilson and McFadden 1996, Ishida et al. 1999). The nucleomorphs, and particularly the genes they contain, have been the Rosetta Stone to understanding secondary endosymbiosis. Chlorarachniophytes are, therefore, a key group for understanding how chloroplasts have spread to many different groups of eukaryotes. Much chlorarachniophyte research has focused on searching for origins of the host and the endosymbiont components (Bhattacharya et al. 1995, McFadden et al. 1995, Van de Peer et al. 1996, Cavalier-Smith and Chao 1997, Ishida et al. 1997, Van de Peer and DeWachter 1997, Keeling et al 1998, Durnford et al. 1999, Ishida et al. 1999, Keeling 2001) in order to dissect who the partners were and what has happened to them in the ensuing amalgamation. A complementary line of enquiry has been to understand the molecular and cellular biology of the nucleomorph genome (McFadden et al. 1994, Gilson and McFadden 1995, Gilson and McFadden 1996, Gilson and McFadden 1997, Gilson et al. 1997, Gilson and McFadden 1998).
Characteristics
All known chlorarachniophytes are unicellular and photosynthetic with green chloroplasts. The group is characterised by the following features:
- Chloroplasts contain chlorophylls a and b.
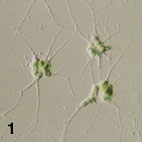
- Amoeboid cells have filose pseudopodia (Fig. 1)
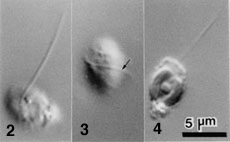
- Flagellated cells bear a single flagellum (Figs 2-4) that wraps around the cell while it swims (Fig. 3).
- Each chloroplast has a prominent projecting pyrenoid, and is surrounded by four envelope membranes (Fig. 5).
- A nucleomorph is present in a space between the second and third envelope membranes of each chloroplast (Fig. 5).
- Thylakoids are often loosely stacked in three.
- The principal storage carbohydrate is beta-1,3-glucan, which is stored in cytoplasmic vesicles that are often tightly associated with pyrenoids (McFadden et al. 1997).
- Mitochondrial cristae are tubular.
Figure 1. Amoeboid cells of Lotharella amoeboformis
extending many filose pseudopodia.
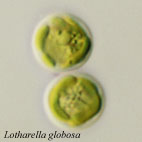
Figure 2-4. Flagellated cells of L. globosa.
Each has a single flagellum that wraps around the cell body when it swims (Arrow). Images copyright © 2001 K. Ishida

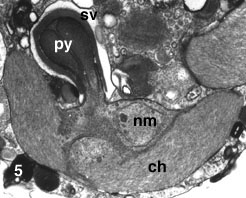
Figure 5. Chloroplast (ch) of L. amoeboformis with a projecting pyrenoid (py) sheathed by a cytoplasmic storage vesicle (sv) that contains beta-1,3 glucan. A nucleomorph (nm) is located in a space between two pairs of chloroplast envelope membranes near the base of the pyrenoid. Image copyright © 2001 K. Ishida
Habitat
Chlorarachniophytes live in warm (tropical to temperate) marine environments, probably throughout the world. Amoeboid and coccoid species have been found from coastal areas, while flagellated members have been found in oceanic waters as picoplankton.
Ultrastructural diversity
Among the chlorarachniophytes, there is a remarkable diversity in ultrastructure of the pyrenoid and localization of the nucleomorph. These traits are used as key characteristics for the generic classification (Ishida et al. 1996).
Life Cycle and Reproduction
The basic life cycle of the chlorarachniophytes comprises amoeboid, coccoid and flagellated cell stages (Fig. 6-C). However, the patterns of the life cycle vary among species, and some species lack one or two of those stages (Fig. 6-A,B) (Ishida and Hara 1994, Ishida et al 1996, Ishida et al. 2000). Coccoid cells in some species are regarded as cysts (Hibberd 1990). Characteristics of life cycle patterns are used to distinguish species in the group (Ishida and Hara 1994, Ishida et al. 2000). Asexual reproduction is carried out by either normal mitotic cell division or zoospore formation. Sexual reproduction has been reported from two species: Chlorarachnion reptans and Cryptochlora perforans. In C. reptans, two different types of cells, amoeboid and coccoid, fuse to form a zygote (anisogamy) (Grell 1990), while in C. perforans, the fusion occurs between two amoeboid cells (isogamy) (Beutrich and Schnetter 1993).

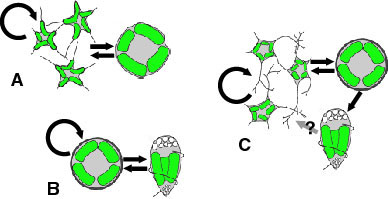
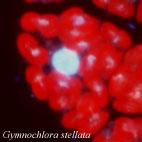
Figure 6. Life cycles of G. stellata that lacks walled and flagellated cell stages (A), L. globosa that lacks an amoeboid cell stage (B), and L. amoeboformis that has all three cell stages (C). Image copyright © 2001 K. Ishida
The nucleomorph
The nucleomorph is a small double membraned organelle located in a space between the second and the third membranes of the chloroplast envelope (Fig. 7). This organelle is the vestigial nucleus of the eukaryotic algal endosymbiont that gave rise to the chloroplasts of chlorarachniophytes (McFadden et al. 1994). The double membranes surrounding this organelle are considered to have originated from the nuclear envelope of the endosymbiont as there are nuclear pore-like gaps in the double envelope (Hibberd and Norris 1984).


Figure 7. A nucleomorph (nm) which is located in a space between the inner (arrowhead) and the outer (double arrowheads) pairs of chloroplast envelope membranes. Nucleomorph envelope has a gap (arrow) that is probably homologous to the nuclear pore of an ordinary nucleus. ch: chloroplast. Image copyright © 2001 K. Ishida
The nucleomorph contains its own genome, which is about 380 kb - the smallest eukaryotic genome known (Gilson and McFadden 1996). The genome consists of three linear chromosomes (Fig. 8), and both ends of each chromosome are capped by telomeres and ribosomal RNA gene cistrons (Gilson and McFadden 1996). It has been estimated that there are about 300 protein genes encoded by this genome (Gilson and McFadden 1997). These genes are arranged in the chromosomes in a highly compacted manner. However, the genes also contain numerous introns, although the introns are extraordinarily small ranging in size from 18 bp to 21bp, smaller than any other spliceosomal introns (Gilson and McFadden 1996). Genes encoded by the nucleomorph genome are mostly 'housekeeping genes', genes for maintaining its replication and expression systems (Gilson and McFadden 1996). Only a few chloroplast-targeted proteins are encoded by the nucleomorph, suggesting that the majority of nuclear-encoded chloroplast genes are now in the host's nuclear genome (Gilson and McFadden 1996).

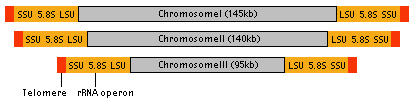
Figure 8. Structure of nucleomorph chromosomes. Image copyright © 2001 K. Ishida
Like other DNA-containing organelles, the nucleomorph is autonomous. Nucleomorph division takes place just before chloroplast division by the sequential infolding of the inner and outer membranes of the nucleomorph envelope (Ludwig and Gibbs 1989). No spindle formation has been observed during the nucleomorph division. How the chromosomes are correctly segregated into two daughter nucleomorphs is still not clear.
Other than in chlorarachniophytes, a nucleomorph is also found in the cryptomonads: flagellates with chloroplasts containing chlorophylls a and c and phycobilins. Despite the striking similarity in structure and genome organization between the two nucleomorphs (Gilson et al. 1998), their origins are clearly different: the cryptomonad nucleomorph came from a red algal endosymbiont, while the chlorarachniophyte one came from a green algal endosymbiont (see Relationships to Other Organisms). Nucleomorphs are thus an excellent example of parallel evolution—the miniaturisation of eukaryotic nuclei to common endpoints (Gilson and McFadden 2001).
Relationships to Other Organisms
Since the chlorarachniophytes arose from a secondary endosymbiosis (Fig. 9), we must consider relationships of the host component and of the endosymbiont to other organisms separately.
All reported molecular phylogenetic analyses (nuclear-encoded small subunit ribosomal RNA gene, tubulin subunits, and actin proteins) on the origin of the host component concur that a heterotrophic amoeboflagellate lineage consisting of the cercomonads and the euglyphinids is the closest relative of the chlorarachniophytes (Bhattacharya et al. 1995, Cavalier-Smith and Chao 1997, Van de Peer et al. 1996, Keeling et al 1998, Ishida et al. 1999, Keeling 2001). However, morphological and ultrastructural data have failed to indicate any particular group of organisms as the closest relative of the host component of the chlorarachniophytes.
For the endosymbiont, all data (including pigment composition and molecular phylogenies) indicate that a green alga was the endosymbiont. However, exactly which green alga was the actual ancestor of the chloroplast is still controversial. Xanthophyll pigment composition suggests that a certain group of the prasinophytes is a possible ancestor of the chloroplast (Sasa et al. 1992). On the other hand, different molecular phylogenetic analyses have suggested alternative candidates: the ulvophytes (Ishida et al.1997, Ishida et al. 1999) or the trebouxiophytes (Van de Peer et al. 1996).

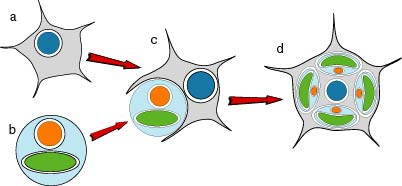
Figure 9. Hypothetical process of the secondary endosymbiotic acquisition of chloroplasts in the chlorarachniophytes. a) an amoeboflagellate eukaryote. b) a green alga. c) engulfment of the green alga by the amoeboflagellate host. d) extant chlorarachniophyte cell containing green chloroplasts each with four envelope membranes and a nucleomorph. Image copyright © 2001 K. Ishida
Cavalier-Smith (1999) proposed the cabozoan theory which states that the chlorarachniophytes and the euglenophytes share a common photosynthetic ancestor that acquired its chloroplast by a secondary endosymbiosis from a green alga. However, molecular phylogeny of the chloroplast EF-Tu (Ishida et al. 1997), and the lack of any observable relationships (morphological or molecular) between chlorarachniophytes and euglenoids lead us to believe that they acquired their chloroplasts separately.
Discussion of Phylogenetic Relationships
The phylogenetic relationships among chlorarachniophyte species have been studied using nuclear- and nucleomorph-encoded SSUrRNA genes (Gilson and McFadden 1998, Ishida et al. 1999). The SSUrRNA gene trees published so far indicate that:
- The chlorarachniophytes are monophyletic.
- The Lotharella and/or Gymnochlora lineages tend to branch out at the base of the chlorarachniophyte lineage.
- Chlorarachnion is monophyletic.
- Flagellated strains form a monophyletic clade.
However, the relationship among major clades is still unclear and the genus Lotharella may not be monophyletic, because Gynmochlora stellata may be closely related to L. amoeboformis (Ishida et al. 1999, Ishida et al. 2000). Futhermore, the phylogenetic position of Cryptochlora perforans is not clear, because neither molecular nor ultrastructural data are yet available.
References
Review Articles
Gilson, P. R., and G. I. McFadden. 1995. The chlorarachniophyte: a cell with two different nuclei and two different telomeres. Chromosoma 103:635-641.
Gilson, P. R., and G. I. McFadden. 1997. Good things in small packages: the tiny genomes of chlorarachniophyte endosymbionts. BioEssays 19:167-173.
Gilson, P. R., U.-G. Maier and G. I. McFadden 1997. Size isn't everything: lessons in genetic miniturisation from nucleomorphs. Curr. Opinion Genet. Dev. 7:800-806
Hibberd, D. J. 1990. Phylum Chlorarachnida. Pp. 288-292 In L. Margulis, J. O. Corliss, M. Melkonian and D. J. Chapman, eds. Handbook of Protoctista. Jones and Bartlett, Boston.
McFadden, G. I., P. R. Gilson, and C. J. Hofmann 1997. Division Chlorarachniophyta. In D. Bhattacharya, ed. Origins of Algae and Their Plastids. pp.175-185. Springer-Verlag Wien New York.
Original Research Articles
Beutlich, A., and R. Schnetter. 1993. The life cycle of Cryptochlora perforans (Chlorarachniophyta). Bot. Acta 106:441-447.
Bhattacharya, D., T. Helmchen, and M. Melkonian. 1995. Molecular evolutionary analyses of nuclear-encoded small subunit ribosomal RNA identify an independent rhizopod lineage containing the Euglyphina and the Chlorarachniophyta. J. Eukaryot. Microbiol. 42:65-69.
Cavalier-Smith, T. 1999. Principles of protein and lipid targeting in secondary symbiogenesis: euglenoid, dinoflagellate, and sporozoan plastid origins and the eukaryote family tree. J. Eukaryot. Microbiol. 46:347-366.
Cavalier-Smith, T., and E. E. Chao. 1997. Sarcomonad ribosomal RNA sequences, rhizopod phylogeny, and the origin of euglyphid amoebae. Arch. Protistenkd. 147:227-236.
Durnford, D. G., J. A. Deane, S. Tan, G. I. McFadden, E. Gantt, and B. R. Green. 1999. A phylogenetic assessment of the eukaryotic light-harvesting antenna proteins, with implications for plastid evolution. J. Mol. Evol. 48:59-68.
Gilson, P. R., and G. I. McFadden. 1996. The miniaturized nuclear genome of a eukaryotic endosymbiont contains genes that overlap, genes that are cotranscribed, and the smallest known spliceosomal introns. Proc. Natl. Acad. Sci. USA 93:7737-7742.
Gilson, P. R., and G. I. McFadden. 1998. Molecular, morphological and phylogenetic characterization of six chlorarachniophyte strains. Phycol. Res. 47:7-20.
Grell, K. G. 1990. Some light microscope observations on Chlorarachnion reptans Geitler. Arch. Protistenkd. 138:271-290.
Hibberd, D. J., and R. E. Norris. 1984. Cytology and ultrastructure of Chlorarachnion reptans (Chlorarachniophyta divisio nova, Chlorarachniophyceae classis nova). J. Phycol. 20:310-330
Ishida K. and Y. Hara 1994. Taxonomic studies on the Chlorarachniophyta. I. Chlorarachnion globosum sp. nov. Phycologia 33(5):351-358
Ishida K., T. Nakayama and Y. Hara 1996. Taxonomic studies on the Chlorarachniophyta. II. Generic delimitation of the chlorarachniophytes and description of Gymnochlora stellata gen. et sp. nov. and Lotharella gen. nov. Phycological Research 44(1):37-45
Ishida K., Y. Cao, M. Hasegawa, N. Okada and Y. Hara 1997. The origin of chlorarachniophyte plastids, as inferred from phylogenetic comparisons of amino acid sequences of EF-Tu. Journal of Molecular Evolution 45:682-68
Ishida K., B. R. Green, T. Cavalier-Smith 1999. Diversification of a Chimaeric Algal Group, the Chlorarachniophytes: Phylogeny of Nuclear and Nucleomorph Small Subunit rRNA Genes. Molecular Biology and Evolution 16(3):321-331
Ishida K., N. Ishida, and Y. Hara 2000. Lotharella amoeboformis sp. nov.: a New Species of Chlorarachniophytes from Japan. Phycological Research 48(4):221-230
Keeling P. J., J. A. Deane, and G. I. McFadden 1998. The phylogenetic position of alpha- and beta-tubulins from the Chlorarachnion host and Cercomonas (Cercozoa). J. Eukaryot. Microbiol. 45:561-570
Keeling P. J. 2001. Foraminifera and Cercozoa are related in actin phylogeny: two orphans find a home? J. Eukaryot. Microbiol. (in press)
Ludwig, M. and S. P. Gibbs 1989. Evidence that the nucleomorphs of Chlorarachnion reptans (Chlorarachniophyceae) are vestigial nuclei: morphology, division and DNA-DAPI fluorescence. J. Phycol 25:385-394.
McFadden, G. I., P. R. Gilson, C. J. B. Hofmann, G. J. Adcock, and U.-G. Maier 1994. Evidence that an amoeba acquired a chloroplast by retaining part of an engulfed eukaryotic alga. Proc. Natl. Acad. Sci. USA 91:3690-3694.
McFadden, G. I., P. R. Gilson, and R. Waller 1995. Molecular phylogeny of chlorarachniophytes based on plastid rRNA and rbcL sequences. Arch. Protistenkd. 145:231-239.
McFadden, G. I., P. R. Gilson, and I. M. Sims 1997. Preliminary characterization of carbohydrate stores from chlorarachniophytes (Division: Chlorarachniophyta). Phycol. Res. 45:145-151.
Sasa, T., Takaichi, S., Hatakeyama, N. and Watanabe, M. M. 1992. A novel carotenoid ester, loroxanthin dodecenoate, from Pyramimonas parkeae (Prasinophyceae) and a chlorarachniophycean alga. Plant Cell Physiol. 33:921-5.
Van de Peer, Y., S. A. Rensing, U.-G. Maier, and R. De Wachter. 1996. Substitution rate calibration of small subunit ribosomal RNA identifies chlorarachniophyte endosymbionts as remnants of green algae. Proc. Natl. Acad. Sci. USA 93:7732-7736.
Van de Peer, Y., and R. De Wachter. 1997. Evolutionary relationships among the eukaryotic crown taxa taking into account site-to-site rate variation in 18S rRNA. J. Mol. Evol. 45:619-630.
About This Page

University of Tsukuba, Tsukuba, Japan
Geoff I. McFadden

University of Melbourne, Victoria, Australia
Correspondence regarding this page should be directed to Ken-ichiro Ishida at
Page copyright © 2009 and Geoff I. McFadden
All Rights Reserved.
- First online 03 August 2001
Citing this page:
Ishida, Ken-ichiro and Geoff I. McFadden. 2001. Chlorarachniophytes. Version 03 August 2001. http://tolweb.org/Chlorarachniophytes/2391/2001.08.03 in The Tree of Life Web Project, http://tolweb.org/








 Go to quick links
Go to quick search
Go to navigation for this section of the ToL site
Go to detailed links for the ToL site
Go to quick links
Go to quick search
Go to navigation for this section of the ToL site
Go to detailed links for the ToL site